기술동향
A new approach to diagnosing the dynamic behavior of DNA and RNA
- 등록일2010-03-15
- 조회수9173
- 분류기술동향
-
자료발간일
2010-03-15
-
출처
RESEARCHSEA
- 원문링크
-
키워드
#DNA#RNA
A new approach to diagnosing the dynamic behavior of DNA and RNA
Molecular fluorescence applied in an unconventional way allows the life-sustaining chemical reactions in genes to be observed in real time
Akimitsu Okamoto
Initiative Research Scientist
Okamoto Initiative Research Unit
RIKEN Advanced Science Institute
The research targets of Initiative Research Scientist Akimitsu Okamoto are nucleic acids; deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). “DNA and RNA form the backbone of life phenomena. For this reason, I want to know when, where and to what extent they act. In other words, I want to diagnose their behavior. I would also like to develop a simple technique that can visualize the s in DNA and RNA dynamically so that we can help diagnose and treat disease.” Conventional DNA and RNA visualization techniques have disadvantages such as low reaction speed and high structural complexity. Gene expression is controlled by methylation, the addition of a methyl group to a base in a DNA sequence called cytosine.
However, it has been very difficult to observe where and to what extent DNA methylation occurs. Utilizing the findings of organic chemistry and photochemistry, Okamoto continued to tackle this tough challenge, and in July 2009, succeeded in observing DNA and RNA in real time, in addition to the technique he developed in 2007 for observing DNA methylation.
Nucleic acids dynamically
DNA is a double-stranded helical structure that consists of four bases: adenine (A), thymine (T), guanine (G) and cytosine (C). The bases A and T, and G and C are always arranged in pairs. Genetic information is encoded as base sequences in the DNA, and the human genome for example comprises as many as three billion of these base pairs.
The human body is made up of about 60 trillion cells and every cell contains DNA with the same base sequence. A gene is a part of a DNA sequence, and the base sequence of the gene is read as RNA. Proteins are produced according to the information in the DNA and are designated to perform various functions. Life could not be sustained if the same gene in every cell functioned at the same time. Instead, the gene is ingeniously activated or d according to the type of cell and the situation. “The quantity and types of RNA in a cell can dynamically according to the activation and suppression of genes. I wanted to observe this dynamic behavior,” says Okamoto. Recently, a research unit lead by Yoshihide Hayashizaki, director of the Omics Science Center at RIKEN, has revealed that there is a lot of ‘non-coding’ RNA without the information necessary to produce proteins. RNAs thus has important functions other than just connecting DNA with proteins.
A protein visualization technique based on fluorescence is now well established. However, for the visualization of DNA and RNA, conventional techniques are slow and structurally complex. Okamoto aimed at devising a simpler and more reliable method. “We brought ideas and technologies based on organic chemistry and photochemistry, my areas of expertise, into biology to develop ‘artificial nucleic acids’, which were the key to achieving the direct observation of DNA and RNA in real time.”
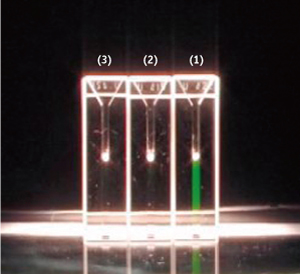
Figure 1. DNA visualization by fluorescing artificial nucleic acid.
Three solutions containing an artificial nucleic acid were mixed with (1) single-stranded DNA complementary to the artificial nucleic acid, (2) single-stranded DNA non-complementary to the artificial nucleic acid, and (3) water. Green fluorescence was produced only in the solution containing single-stranded DNA complementary to the artificial nucleic acid (1).
Diagnosing DNA and RNA using 12 colors of fluorescent dye
“It is simple. I will do it for you here,” says Okamoto, showing his experimental setup. He has three glass vials of a solution containing an artificial nucleic acid. The solution is light orange, which is the color applied to the artificial nucleic acid. To one of these three vials Okamoto adds single-stranded DNA having a complementary base sequence that can bind to the artificial nucleic acid; to the next he adds single-stranded DNA having a non-complementary base sequence; and to the last vial he adds plain water. After stirring gently, the solution containing the complementary DNA rapidly s to fluoresce a bright green (Fig. 1). “We arranged things so that green fluorescence is produced only when the artificial nucleic acid couples with the DNA,” says Okamoto. No special operation is needed other than mixing the solutions at ambient temperature, and no special equipment is needed. The solution can produce fluorescence under the light of an incandescent lamp, and can be discriminated by the naked eye.
What was the key that enabled Okamoto to visualize DNA or RNA, which had presented so much difficulty? “It is the fact that we took into account a phenomenon called ‘excitonic interaction, which had not been considered in biology,” he replied. excitonic interaction is a phenomenon by which the emission from a fluorescent dye molecule is quenched when the dye molecules stack parallel to one another. When the dye molecules are isolated, the fluorescence becomes observable. From among the dyes that exhibit excitonic interaction, Okamoto ed on thiazole orange (Fig. 2). “We created an artificial nucleic acid, having base sequences complementary to the target DNA or RNA, and coupled one of the base with two thiazole orange molecules, holding them stacked in parallel. The artificial nucleic acid does not produce fluorescence unless it reacts with a single-stranded DNA or RNA complementary to the nucleic acid, because the dye molecules remain in parallel. When the artificial nucleic acid reacts with complementary single-stranded DNA or RNA, however, the dye molecules are separated among the base pairs and to fluoresce individually.”
A commercial automatic DNA/RNA synthesizer is used to synthesize the artificial nucleic acids. The instrument is capable of automatically synthesizing artificial nucleic acids consisting of up to 100 bases from samples of the artificial nucleotides on the basis of the defined base sequence.
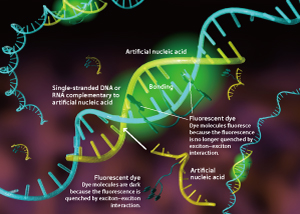
Figure 2. Artificial nucleic acid that fluoresces when it binds to a target nucleic acid.
An artificial nucleic acid having base sequences complementary to the target DNA or RNA is synthesized and modified by adding a couple of dye molecules (e.g. thiazole orange) to one of the base sequences. In the absence of single-stranded DNA or RNA complementary to the artificial nucleic acid, the dye molecules remain ‘dark’ due to excitonic interaction. In the presence of single-stranded DNA or RNA complementary to the artificial nucleic acid, however, each dye molecule can fluoresce because of the isolation of the dye molecules by the interaction with DNA or RNA nucleotides.
Conventional DNA or RNA visualization techniques also have another serious drawback in that the fluorescence remains active even after the RNA sequence has been disintegrated. “In our method, no fluorescence is produced when the DNA or RNA connecting the artificial nucleic acid has been disintegrated because the dye moieties are arranged in parallel again. The quantity of DNA or RNA can be determined by measurement of fluorescence intensity.”
Dyes other than thiazole orange also exhibit excitonic interaction. Okamoto’s research unit has already created an artificial nucleic acid with one of 12 colored dyes attached to it (Fig. 3). This artificial nucleic acid can be easily introduced into living cells. “We can observe a series of processes in real time, in which an RNA molecule comes out from the nucleus of a cell and spreads out into the cell. The RNA molecule functions then in translation process of a protein synthesis and later disintegrated. If we use artificial nucleic acids containing different colored dyes, we can investigate various RNA and DNA behaviors at the same time. Also, it has been recently found that micro-RNA, small RNA with about 20?25 bases, regulates the gene expression through a mechanism called ‘RNA interference’. Micro-RNA is so small in size and quantity that it is very difficult to visualize. However, I really want to find a way to observe it. Thus, we are developing a technique that can enhance the fluorescence intensity by more than 100 times.”
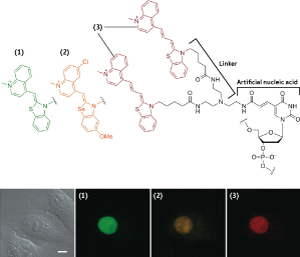
Figure 3. Structure of fluorescing artificial nucleic acid.
A base, (uracil in this figure, an RNA base), of an artificial nucleic acid is modified by a linker (additive part). A pair of dye molecules (3) is then added at the end of the linker. Using different dye molecules ((1) or (2)) can produce different colors. Three of the 12 colored artificial nucleic acids developed by the Okamoto research unit are shown in this figure. The lower photos are examples of intracellular multi-color live cell RNA imaging. Three kinds of artificial nucleic acid were injected into the nucleus of a cell injected with three types of micro-RNA, allowing the three kinds of micro-RNA to be discriminated by fluorescent colors simultaneously.
Discriminating a single-base difference, leading to medical care assistance
“Dr Hayashizaki, director of the Omics Science Center at RIKEN, is now attempting to use the artificial nucleic acids we developed in clinical practice,” says Okamoto. The DNA base sequences of all humans are almost exactly the same, since we belong to the same species. However, some portions differ slightly among individuals. These differences are called ‘gene polymorphisms’. Single nucleotide polymorphism (SNP) is the term used to refer to the difference on a single base. There is about one SNP in every few hundred bases, and such differences are known to be related to differences in susceptibility to disease, efficacy of drugs and the occurrence of adverse effects. If the SNPs of patients can be examined as part of clinical practice, the knowledge obtained will lead to personalized medicine, administering of the drug with the highest efficacy and least adverse effects for each individual patient. SNP-based diagnosis, however, is not popular because it requires extensive specialty equipment, and several hours to several days to obtain results.
Hayashizaki developed the SmartAmp method in 2007 with the aim of simple, rapid and accurate SNP diagnosis. The SmartAmp method is a technique in which multiple enzymes are used to ively amplify the DNA containing the target SNP and then diagnose the presence of the SNP from a blood sample. “Dr Hayashizaki ed on the artificial nucleic acids we had developed during his efforts to refine his SmartAmp method. In his new SmartAmp method, when a reaction is initiated between the blood and the enzymes, an artificial nucleic acid with a base sequence complementary to the target SNP is added. Any target SNPs present multiply together with the artificial nucleic acid attached to them, causing the specimen to produce fluorescence after the reaction. If there is no target SNP, no fluorescence is produced. This SmartAmp method allows us to diagnose the presence of SNPs simply and reliably because the fluorescent dye can produce fluorescence dozens of times brighter than with the conventional SmartAmp method.” Only a single drop of blood is required, and the process generally takes only 30 minutes to yield results.
Okamoto points out, “In most current SNP diagnoses, all base sequences are determined. The information, however, should be dealt with carefully because these base sequences are the ultimate personal information. The SmartAmp method is only used to examine the presence of target SNPs. It is extremely important to obtain only the necessary information.” Practical applications of SmartAmp are also being developed for the detection of new flu viruses.
Observing a single methyl group binding to a DNA sequence
The Okamoto Initiative Research Unit is also developing another visualization technique. “Research subjects on nucleic acids are schematized to be a pyramidal structure with four stories,” says Okamoto. From the bottom, the hierarchy consists of the gene, the DNA base sequence, SNPs and epigenetics. “Roughly speaking, research into nucleic acids has advanced in this order. The higher the story, the smaller the size of the target and the harder the research subject.”
In the visualization of DNA methylation, Okamoto is working on epigenetics, at the top of the research pyramid. Epigenetics is the study of processes in which gene expression is controlled by chemical modifications to DNA without changing the base sequences. Epigenetics includes DNA methylation, acetylation and methylation of histones wrapped by DNA, as well as phosphorylation. “DNA methylation is the process by which a methyl group is attached to cytosine in DNA sequence. When the cytosine is methylated, gene expression is inhibited. DNA is always regulated by methylation and the other modification. I want to observe this behavior, but it is extremely difficult to find a single methyl group attached to the ‘giant’ DNA molecules. We need an atomic-level precision approach.”
In 2007, Okamoto succeeded in developing a system capable of detecting DNA methylation based on the ‘ICON’ probe, an artificial nucleic acid modified by a substituent with bipyridine and metallic osmium. “It is easy to understand the principle behind my ICON method if you compare it to fishing,” says Okamoto (Fig. 4). The river can be compared to the giant DNA, the fish to the methylated DNA, and the ICON probe to the fishing rod. An artificial nucleic acid with a base sequence complementary to the DNA methylation aims the ‘cast’ of the fishing rod, while the bipyridine, an organic molecule, acts as the ‘hook’ and the osmium acts as the ‘bait’. Once casted, the angler needs simply to wait to catch methylated DNA site when it occurs.
“We use a metal complex reaction,” explains Okamoto. “Osmium forms a metal complex that can oxidize double bonds. A non-methylated cytosine has two double bonds, whereas a methylated cytosine has three double bonds. Since the methylated cytosine has a larger number of double bonds than the non-methylated cytosine, it complexes with osmium 400 times faster than the non-methylated cytosine. Thus, we utilize the difference in the reaction rate to detect DNA methylation.” Bipyridine is essential for the formation of a complex, and promotes the oxidation.

Figure 4. Principle of ICON, a methylated DNA analysis system.
ICON consists of an artificial nucleic acid, the organic molecule bipyridine and metallic osmium. The ICON method involves a chemical reaction in which the osmium oxidizes the methylated cytosine to form a metallic complex, thus catching methylated DNA. The sequence of the artificial nucleic acid is complementary to the region to be examined for the presence of any methylation.
Conventionally, methylation has been detected by the restriction enzyme method and the bisulfite method. However, these methods take several to a dozen or more hours to complete, and the DNA sequences are chopped into small fragments. In contrast, the reaction for the ICON method occurs in just 10 minutes to an hour at ambient temperature. The ICON method also detects methylated DNA without breaking the DNA into fragments. An instrument for polymerase chain reaction (PCR) can be used to multiply the methylated DNA to determine the amount of methylated DNA. “The ICON method is the first method in the world to make it possible to detect whether or not a single cytosine in a giant DNA molecule is methylated.”
Diagnosing methylation for early cancer diagnosis
Okamoto has received many enquiries about the ICON method both from Japan and internationally. There are reasons for this worldwide attention. “It is considered that abnormalities in the location and volume of methylated DNA are closely related to the occurrence of cancers and aging. Epigenetics research related to methylated DNA just ed. However, I am expecting that DNA methylation can be used as an indicator for early cancer diagnosis and treatment strategies in the future once the relationship between methylated DNA and diseases has been clarified.” The ICON molecules were commercialized by Gene Design in 2007. “I will be pleased if the ICON method is used by as many people as possible, and linking the methylated DNA detection technique to clinical practice will contribute to its development.”
Okamoto continues, “Developing a method and writing a paper is not our goal. I think we will receive recognition when the newly developed techniques are used by many people. We are always conscious of contributing to medical care. That is the reason why we used the term ‘diagnose’ in the titles of our papers and presentations.” Techniques developed by the Okamoto Initiative Research Unit will be actively used in clinical practice in the near future.
About the Researcher
Akimitsu Okamoto
Akimitsu Okamoto was born in Nagoya, Japan, in 1970. He graduated from the Faculty of Engineering, Kyoto University, in 1993, and obtained his PhD in 1998 from the same university. After a year of postdoctoral researcher at the Department of Chemistry, Massachusetts Institute of Technology, USA, he returned to Japan as a research associate at the Department of Synthetic Chemistry and Biological Chemistry, Faculty of Engineering, Kyoto University, where he ed his career in photochemistry and synthetic nucleic acids chemistry. He moved to the RIKEN Frontier Research System, now the RIKEN Advanced Science Institute, as an initiative research scientist in 2006. Since then, he has acted as unit leader of the Okamoto Initiative Research Unit. His research es on the design, synthesis and physical properties of new artificial biopolymers with various functions and the design of new organic chemical systems for recognizing, transforming and visualizing a single component or atom in biopolymers of interest.
지식
- BioINwatch 어떻게 하면 단백질 구조 만큼 RNA 구조도 정확히 예측할 수 있을까? 2025-04-03
- BioINwatch 후성 유전체 비트(Epi-bits)를 활용한 DNA 기반의 데이터 저장 기술 2025-03-18
- BioINwatch RNA 구조체 분석을 통해 규명한 단백질 번역 조절의 새로운 메커니즘 2024-12-05
- BioINwatch DNA 컴퓨팅, 미래의 데이터 처리와 저장 기술 2024-11-26
- BioINwatch '중심 원리(Central Dogma)' 패러다임을 확장한 microRNA 2024-11-18
동향

