기술동향
Exploring biological phenomena using the world’s fastest computer system
- 등록일2008-08-08
- 조회수5984
- 분류기술동향
-
자료발간일
2008-07-25
-
출처
RIKEN
-
원문링크
-
키워드
#시스템생물학#System biology
출처 : RIKEN
Exploring biological phenomena using the world’s fastest
computer system
Makoto Taiji
Deputy Project Director
High-performance Molecular Simulation Team
Computational and Experimental System Biology Group
Genomic Sciences Center
The fastest special-purpose computer system for molecular simulations in the world is available at the RIKEN Genomic Sciences Center (GSC). Developed by Makoto Taiji, team leader of the High-Performance Molecular Simulation Team, Computational and Experimental Systems Biology Group in GSC, the MDGRAPE-3 system is capable of calculating interatomic forces and simulating their motions. As such, MDGRAPE-3 is expected to make significant contributions to faster development of new drugs with higher efficacy, and the understanding of biological molecules, such as proteins, at the atomic level. The following is an interview with Taiji about the prospects for life sciences explored with MDGRAPE-3, and the secret story of its development.
MDGRAPE-3, the world’s fastest computer system
When the elevator door opens, a booming sound can be heard everywhere on the fifth floor of the West Building in the RIKEN Yokohama Institute. It comes from the room in which the MDGRAPE-3 is installed (left panel in Fig. 1).
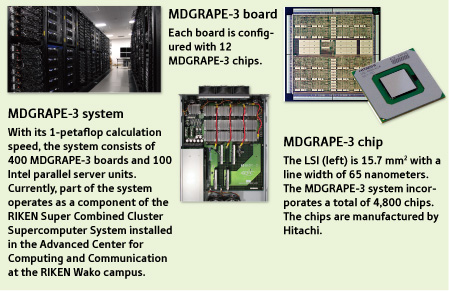
Figure 1: MDGRAPE-3 special-purpose computer system for molecular dynamics simulations.
In June 2006, the MDGRAPE-3 system marked the world’s highest speed of 1 petaflop. The petaflop is a unit of measurement for the processing speed of a computer system; each petaflop is equivalent to 1015 (one quadrillion) floating point operations per second. MDGRAPE-3 is capable of doing one quadrillion calculations per second. This performance is about 20,000 times faster than the latest personal computers. “Even now MDGRAPE-3 remains the only computer system that has achieved a calculation speed of 1 petaflop,” says Taiji, the developer of the system.
To understand biological phenomena at the atomic level using a computer system is the goal of the High-Performance Molecular Simulation Team led by Taiji. “It is our policy to undertake a full range of work, from hardware production to software development, and applied research,” he says. “There are almost no other research groups like us worldwide.”
Personal computers in routine use are ‘general-purpose’ computers, capable of doing a broad range of different types of calculations.
The MDGRAPE-3 system comprises a combination of general-purpose computers and special-purpose computers dedicated to calculating the forces exerted between atoms that constitute materials. Chemical binding forces, electrostatic forces, and intermolecular forces are exerted on atoms, which move according to equations of motion that describe the relation between these forces and the response of the atoms. Because any material comprises a vast number of atoms, the forces exerted on individual atoms cannot be calculated to simulate their motions without powerful computers.
Taiji characterizes the MDGRAPE-3 system by three aspects. “First,” he explains, “MDGRAPE-3 became the first computer in the world to achieve a 1-petaflop calculation speed. Second, the cost performance is high. Our system has been developed with about one billion yen, which is only a tenth of the money spent in developing an ordinary general-purpose large computer system used at a university or research institute. Third, the power consumption is small.”
The key to his success in achieving the world’s fastest processing speed lies in the use of broadcast-based memory architecture. A great many calculations can be done at one time because each memory device contains multiple computing units connected in parallel. In the MDGRAPE-3 system, as many as 720 calculations can be performed simultaneously with one ‘LSI’-an ultrafine electronic circuit mounted on a semiconductor substrate that serves as the core of a computer system. The number of calculations per LSI is limited to around 16 with the use of a current ordinary general-purpose processor.
The amount of heat generated per unit surface area by the latest processor is as much as that on the surface of a nuclear reactor containment vessel at a nuclear power station. It is critical to minimize power consumption by reducing this heat. The power consumption of MDGRAPE-3 is 200 kilowatts per hour, one hundredth the level of a general-purpose large computer system. Even so, a huge cooling system is required, and it is this that produces the loud booming sound on the floor where MDGRAPE-3 is installed.
Faster development of new high-efficacy drugs
The development of the MDGRAPE-3 system began in 2002 as part of the Protein 3000 Project, Japan’s national project to explore protein structures and functions. “The structures and functions of many proteins have been elucidated. Now importance must be given to handling the huge amounts of information obtained,” points out Taiji. “For this, MDGRAPE-3 was necessary.”
One expectation of the MDGRAPE-3 system is in the development of new drugs. Proteins have pockets known as active sites, to which particular molecules bind to allow the proteins to function. If the structure of a disease-related protein is clarified, it will be possible to find molecules that bind to the active sites of the protein efficiently, and to design molecular shapes that facilitate the binding.
Each protein has more than a thousand drug candidate molecules. The conventional approach to ing the most suitable molecule comprises simulations by the docking method. Candidate molecules are applied to protein active sites one by one, and the molecules that exactly fit are ed. However, Taiji points out a problem with the docking method. “In the docking method, proteins are handled as solid matter with fixed shapes, and the handling of water, the most important molecule for all organisms, is inadequate.” He explains that proteins in action in the body are constantly oscillating in the presence of thermal energy. “Even if a candidate molecule is found to exactly fit an active site, this does not always mean high affinity, and the effect of water must be considered in determining the action of the molecule.”
MDGRAPE-3 is capable of simulating the dynamics of proteins and candidate molecules, including water, at the atomic level, to enable the accurate detection of binding molecules. “Simulations are often used to understand functions,” says Taiji. “And my goal is to predict functions, and to control those functions by designing molecular shapes.”
A protein comprises a sequence of amino acids arranged according to genetic information. It cannot function normally unless the amino acid sequence is accurately folded. However, much remains unknown about the way in which the protein is folded. Taiji and his colleagues have succeeded in simulating the folding of Chignolin, the world’s smallest artificial protein, which consists of only 10 amino acid residues (Fig. 2). The protein in the elongated state bends and elongates repeatedly while oscillating, until it gradually reaches the right shape. This represents a major step toward the functional elucidation of protein folding.
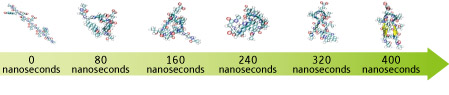
Figure 2: Simulation of the protein folding process.
Chignolin is a small protein consisting of 10 amino acid residues. Simulated is the process by which the protein is three-dimensionally folded from the state of amino acids in sequence. Each particle indicates an atom. Although water molecules do not appear here, their motions were also included in the calculations.
The MDGRAPE-3 system won the Gordon Bell Prize Honorable Mention (Peak Performance division) in 2006. This world-class commendation for the performance of high-speed computers is sponsored by the Association for Computing Machinery. “As a researcher engaged in developing computer systems, I am very happy to receive the prize,” says Taiji with a smile. His winning article concerns simulations of the process by which peptides from the yeast protein Sup35 aggregate in water to form needle-like crystals (Fig. 3). The motions of 17 million atoms, including the surrounding water, were calculated. “Protein aggregation is involved in the onset of nervous diseases such as Alzheimer’s disease and Parkinson’s disease,” he adds. “I am working with the expectation that my work will help elucidate the mechanism for the onset of nervous diseases.”
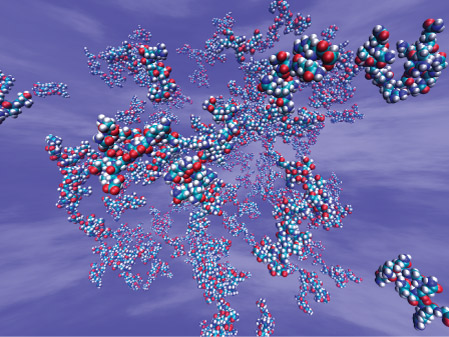
Figure 3: Simulation of the protein aggregation process.
The process by which part of the yeast protein Sup35 aggregates in water to form needle-like crystals is simulated. Although water molecules are not shown here, the motions of 17 million atoms, including water, were calculated (background cloud is an image).
The secret story of the development of MDGRAPE-3
“I was optimistic that it would go well,” says Taiji, looking back on the early days when he began developing the MDGRAPE-3 system. However, the reality was quite different.
Development of a computer system begins with the design of the LSIs. “I devoted myself to sitting in front of a computer day after day writing LSI programs,” says Taiji in retrospect. “That year was very hard. At one time, because I was always thinking in computer language, I found myself unable to talk with my family when I returned home from the office.”
The electron travels at a speed of 200,000 kilometers per second-two thirds the velocity of light. “As the circuit operating speed has been increasingly getting faster, the speed of the electron must now be taken into account when designing LSIs.” The LSI for MDGRAPE-3 measures 15.7 x 15.7 mm2 (right panel in Fig. 1). Taiji says that sometimes he had to keep in mind the speed of electrons moving from one end of the LSI to the other.
In December 2003, the design of the LSI was completed. Taiji proceeded to prepare the MDGRAPE-3 board with the LSI mounted on it, and attempted to check its performance. At this stage, however, a problem arose. “The chip did not work well despite my efforts.” Although the accuracy of the circuit connections and the normal performance of the chip had been verified by computer, the chip was failing. Redesigning the LSI would cost around an additional one hundred million yen. “At that time, I noticed my heart beating and my blood pressure rising,” adds Taiji. Several days later, the fault was located, not in the chip, but in the software, and the project proceeded smoothly.
The MDGRAPE-3 chip (right panel in Fig. 1) is capable of calculations at 230 gigaflops. This performance is more than 30 times higher than the then fastest Intel Pentium 4 chip, and boasts the world’s highest performance per single chip.
Twelve of the MDGRAPE-3 chips were mounted on a board (middle panel in Fig. 1). At that stage, many problems arose. Budget cuts, also brought trouble, but the 1 petaflop speed was achieved in June 2006. “All my efforts became worthwhile,” says Taiji. Capable of 1 petaflop calculations, the MDGRAPE-3 system is configured with 400 boards carrying the MDGRAPE-3 chips and 100 Intel server units. Special-purpose computers and general-purpose computers are combined, as with the first computer system Taiji constructed. “People of an earlier generation might think about how to construct the system as a whole,” he says. “But I grew up in the age of personal computers. Since I had been fond of building radios and electronic gadgets since my childhood, I began constructing a computer system with a light-hearted curiosity?to add something to the existing personal computer.”
The first computer system Taiji constructed was a special-purpose machine for simulating a magnet known as m-TIS in 1987, when he was a graduate student. Later, he joined the development project for GRAPE, the special-purpose computer system for astronomical simulations at the University of Tokyo. GRAPE is capable of calculating gravitational forces between stars to simulate the motions of celestial bodies. By replacing stars with atoms and gravitational forces with electromagnetic forces and intermolecular forces, MDGRAPE-3 was born.
When he was a graduate student, Taiji specialized in laser spectros. His doctoral thesis concerned measurement of s in Rhodopsin, a protein in the eye, when exposed to light. Although constructing computer systems was no more than a hobby, he entered the field of computer systems when about to leave graduate school. At present, he is challenging the simulation of proteins using the world’s fastest computer system, which he developed himself. Twenty years have elapsed since he built his first computer. “I am surprised at what I have achieved. In those days, 1 petaflop calculations were nowhere in my mind.”
The problem concerns time
“Our next goal is to extend the simulation time,” says Taiji. Binding to other molecules, folding, and other phenomena in proteins occur on timescales of microseconds (10?6 seconds, that is, one millionth of a second) or above. They cannot be simulated unless calculations are made at intervals of one femtosecond (10?15 seconds, that is, one quadrillionth of a second) in accordance with the speed at which protein atoms move, and the results are integrated from these calculations. A large gap of nine orders of magnitude exists between a femtosecond and a microsecond. Even with the use of MDGRAPE-3, it takes one day to simulate a microsecond phenomenon. One possible solution to this problem may be to increase the computing speed, but this is subject to limitations. Taiji says that he is thinking of another approach. “I am going to perform multiple short-timescale simulations and join them together, so I can watch long-timescale phenomena.”
We asked him about any future plans for MDGRAPE-4. “Yes, I am planning to construct a general-purpose computer system,” answers Taiji. His plans include the newly introduced concept of a ‘tile processor’, which comprises LSI-mounted chips arranged like tiles. The planned system is expected to perform at about 10 times higher speeds than existing systems while ensuring general-purpose quality with the tile arrangement, and completion is scheduled in four years. “Computer systems are experimental apparatus. I want to construct a more sophisticated system that will beat all the others.”
About the researcher
Makoto Taiji was born in Tokyo, Japan, in 1964. He graduated from the Department of Physics, Faculty of Sciences, the University of Tokyo, in 1986, and obtained his PhD in 1991 from the same university. From 1991 to 1996, he was the assistant professor of the Graduate School of Arts and Sciences at the University of Tokyo. From 1997 to 2002, he was the associate professor of the Institute of Statistical Mathematics. He joined the Genomic Sciences Center, RIKEN, as a team leader and was promoted to deputy project director in 2007. Currently he is the group director of the computational systems biology research group at the RIKEN Advanced Science Institute. His research es on the applications of high-performance computing in life sciences, and dedicated computer architectures for scientific simulations. He was awarded the Gordon Bell Prize in 1995 for developing a ‘Special-Purpose Machine’, and in 2006 he received an Honorable Mention for the same award for the ‘Peak Performance’ achieved by the computer he developed.
...................(계속)
☞ 자세한 내용은 내용바로가기 또는 첨부파일을 이용하시기 바랍니다.
관련정보
동향
- 기술동향 2019 Keystone Symposia: Tumor Metabolism 참관기 2019-04-29
- 기술동향 대사과정에 대한 시스템생물학 2017-07-06
- 기술동향 System biology 동향: Protein-protein interaction network 구축현황과 활용 2017-03-29
- 기술동향 시스템생물학의 합성생물학 적용 연구동향 2015-09-15
- 정책동향 Strategic Report for Translational Systems Biology and Bioinformatics in the European Union 2012-02-14

